PROFESSOR AHNA SKOP
There are several large gaps in how we teach science today that I have tried to address: 1) How do we get students to understand key concepts, 2) have them apply the knowledge they learned, 3) give them skills to communicate science effectively. You can know the facts, but if you can’t apply them or communicate them well, how can you convince others that your ideas are important? With our current political and funding climate, these communication skills are increasingly critical for science policy that affects us all. To address these issues, I created a 100% active learning course that enables them to think like a scientist and learn to communicate more effectively. In my course, students ask questions, participate in discussions, interact in groups, write Specific Aims, come up with hypotheses, experience peer review, and learn how to present their work visually and orally on a website they build and a final presentation at the end of the semester. In turn, these experiences have made students more confident asking questions, forming hypotheses, and what it takes to be a scientist in today’s world.

Creating an inclusive learning environment using Growth Mindset Theory: There are many ways of knowing and means of learning, but creating an inclusive learning environment is key to student success regardless of background or ability. The first way I create a positive teaching and learning climate is first getting to know my students (by first name/hometown/hobbies/passions), introducing how similar I am to them, and sharing my path in science. The second way I do this, is by grading backwards. Here, I give all of the students the total points on day one and subtract points over the course of the semester. This is based on the Growth Mindset Theory. Here, the total points at the beginning of the semester allows the students to develop their knowledge and skills as a scientist. Being a scientist is not something you are born with but can be developed. Losing points is a positive as a few points here and there doesn’t create major stress for the students. ‘A’ students and ‘C’ students are treated as equals on day one. All students accept the challenge to the semester long project and the outcomes are remarkable. (see student project websites: genetics564.weebly.com/projects)
Grading:
The total points possible is 800. Everyone starts with an “A” (800pts) on day one and then you lose points over the course of the semester. Everyone has the potential to get an “A” in this course. It is impossible to fail unless you don’t do any work or participate in class.
CREATING A WEB-BASED PROJECT ON A GENE AND DISEASE
The goal of this project is an in-depth, genomic and bioinformatic analysis of a human disease gene or trait. This disease gene must be homologous to one or more genes in one or more model organisms (the bacterium E. coli, the single-celled yeast, S. cerevisiae, the budding yeast, S. pombe, the roundworm C. elegans, the arthropod D. melanogaster, the mouse M. musculus, and the plant, Arabidopsis thaliana).
Keep in mind that your project should include both a “global” analysis of your selected disease gene and the implied “local” analysis of a single, isolated gene. A global analysis reflects the central themes of the course: structural, functional (both transcriptomics and proteomics), and comparative genomics. A local analysis reflects the gene’s structure and function and its primary role in the disease etiology. Both analysis types should reflect the intricate relationships among sequence, structure, function, genome organization and expression, and evolution. The lab activities throughout the semester will assist you in conducting these analyses.
You will, of course, be making extensive use of bioinformatics programs and databases found on the Internet (NCBI, etc.). My evaluation of your project will be based in part on the depth of your analyses, and on how well you use these available bioinformatics tools. But a tool is only a tool, and its use generates data that must be interpreted, analyzed, compared, and critiqued from a variety of perspectives. Finally, conclusions must be drawn along with recommendations for future experiments or analyses.
The format of your project should be that of a Web Page, so you will want to use Weebly.com (or any other web-based program you are familiar with) to construct the final version with all text and images. The length depends on the amount of information associated with a particular disease gene, and you should strive for brevity and clarity.
• Find a disease or trait and an associated gene/protein for which they have found a mutation.
• You will be working on this gene/protein for the entire semester.
• You will craft a 1-page Specific Aims that you will work on the entire semester
• You will present your FINAL Specific Aims at the end of the semester in a 15-minute presentation to the class.
• You will have built a website with all of the research and bioinformatic information you have obtained in class, outside class and in lab. This website will be your published work which can be used in your resume as published research.
• Your Specific Aims, final talk and website will be peer reviewed by the class and by the instructor.
Gen 564 Semester-Long Project Details
Select a human genetic disorder or trait, by which we mean a disease or heritable predisposition to a certain condition. It can be a single gene disease/trait, or a polygenic one. In the case of a polygenic disorder, you need only do the genomic analysis for one of the involved genes. The role of the selected gene in disease should be featured in your project, and you should review the underlying genetic evidence for the role of the gene in the disease. Some disorders are not polygenic traits, but you will find that more than one gene can cause or predispose different individuals to the disease (BRCA1 and BRCA2 for heritable predisposition to breast cancer, for example). In a case like this, you would introduce both genes as causing the predisposition, and then select one to focus on in detail for the rest of the website project.
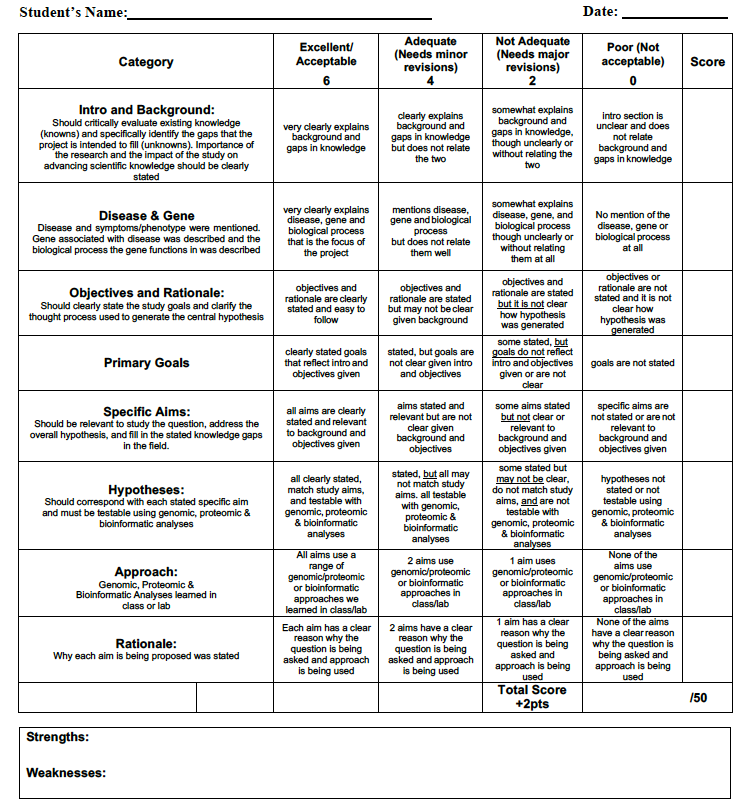
Specific Aims: Over the course of this semester you will be writing a Specific Aims for your project to help you craft a hypothesis and experimental approach for your project. You must use genomic and proteomic techniques you learned in class that would address the specific aims you are proposing and discuss why these techniques are appropriate for your project. You should have 3 aims in your Specific Aims. Well-defined specific aims or objectives are key to the successful completion of a research project. The specific aims should answer the question “What are you going to do and why?” You will need to spend more time planning than writing. You really need to think, read and do research about your project before you proceed until you can ASK a Question! Then: 1) Decide on a hypothesis, 2) Define the aims to test your hypothesis and 3) Choose genomic and/or proteomic approaches that you learned from your readings that you think are the best way to address your hypotheses and questions. Mapping out a strategy will pay off in the end. HINT: Each week your readings are chosen to be models for genomic and proteomic approaches for your own project. So pay attention and get inspired by your readings and your research. By writing specific aims, you will be prepared for your final presentation and in writing up your conclusions and future research sections of your project. Your peers and Ahna will be evaluating you with rubrics over the course of the semester, just as you would experience as a scientist in real life.
Hypothesis: Your hypothesis triggers everything you do. Conceptually, think of it as your destination, determining the course of your semester-long research. Choose a hypothesis that is well focused and testable, and that your experimental results will be able to prove or disprove. Your classmates and Ahna must believe that your hypothesis is sound and important enough to be tested.
Final Presentations: You are going to present what you learned and found out about your gene/protein in the last few weeks of class. This presentation is 15 minutes long. It’s about a 12-minute talk with 3 minutes of questions. Briefly go over what your main findings over the course of semester were for your gene/protein and how these findings helped you craft your hypothesis (which you will state) and the subsequent experiments you are going to perform to test these hypotheses.
Conclude with a brief discussion of what you think is the most important thing to find out next about this disorder/trait and the gene(s) that cause it, and discuss why. You should assume that lab money and manpower are not your limitations.
Presentation rough draft: Successful final presentations result from well-planned and thought-out presentation drafts. You will turn in 2 rough drafts of your final presentations. Ahna will provide you feedback and a time to meet with her one-on-one.
Final Website: Your final web project includes all of the assignments plus data that you obtained about your gene/protein over the course of the semester. Using bioinformatic approaches, you are to use what you learned from our in-class discussions and in-lab hands-on experiences about a particular technique and online database to figure out everything there is to know about your gene/protein. By the end of the semester you should have discovered a lot. And by all means you are welcome to venture off to any online database that is available to you to try to understand and possibly come up with some ideas on how to “cure” your disease that you are studying. The website is not due until after finals and can will be considered published work by you! You can use this on your resume as published research.
References: Put the references to journal articles, images and websites (including bioinformatic websites you use) that you used over the course of the semester on your project on each page that you create. Figures should be referenced as you use them on each page in a figure legend. Every page that you create should have references (overall grade by-10% pts if this is not the case). If you use a bioinformatic website, like PFAM, hyperlink out to that website you used for each analysis. *References should be on every slide where you have shown data or images that are not your own
FINAL Specific Aims Rubric